FGF23 (C-terminal) multi-matrix ELISA
-
Category number
BI-20702
-
Method
Sandwich ELISA, HRP/TMB, 12×8-well detachable strips
-
Sample type
Serum, EDTA plasma, citrate plasma, heparin plasma
-
Sample volume
50 µl / well
-
Assay time
20-24 h / 1 h / 30 min
-
Sensitivity
0.08 pmol/l (= 0.6 pg/ml)
-
Standard range
0 – 20 pmol/l (= 0 – 154 pg/ml)
-
Conversion factor
1 pg/ml = 0.133 pmol/l (MW: 7.52 kDa)
-
Specificity
Human FGF23 (Intact and C-terminal fragments of endogenous and recombinant human FGF23)
-
Precision
In-between (n=10): ≤ 10 % CV
Within-run (n=6): ≤ 12 % CV
-
Use
CE marked – for IVD use in the EU
-
Validation Data
See validation data tab for: precision, accuracy, dilution linearity, values for healthy donors, etc
Product Overview
The FGF23 (C-terminal) immunoassay is an overnight, 96-well sandwich ELISA for the quantitative determination of FGF23 in human serum and plasma. The assay employs human serum-based standards to ensure the measurement of biologically reliable data.
The FGF23 (C-terminal) kit uses antibodies with characterized binding affinities.
Principle Of The Assay
The FGF23 (C-terminal) ELISA kit is a sandwich enzyme immunoassay for the quantitative determination of FGF23 in human serum and plasma samples.
Figure explaining the principle of a sandwich ELISA:
In a first step, standard/control/sample and detection antibody (biotinylated polyclonal rabbit anti-human FGF23) are pipetted into the wells of the microtiter strips, which are pre-coated with anti-human FGF23 antibody. FGF23 present in the standard/control/sample binds to the pre-coated antibody in the well and forms a sandwich with the detection antibody. In the washing step all non-specific unbound material is removed. In a second step, the conjugate (streptavidin-HRP) is pipetted into the wells and reacts with the detection antibody. After another washing step, the substrate (TMB, tetramethylbenzidine) is pipetted into the wells. The enzyme-catalyzed color change of the substrate is directly proportional to the amount of FGF23. This color change is detectable with a standard microtiter plate reader. A dose response curve of the absorbance (optical density, OD at 450 nm) vs. standard concentration is generated, using the values obtained from the standard. The concentration of FGF23 in the sample is determined directly from the dose-response curve.
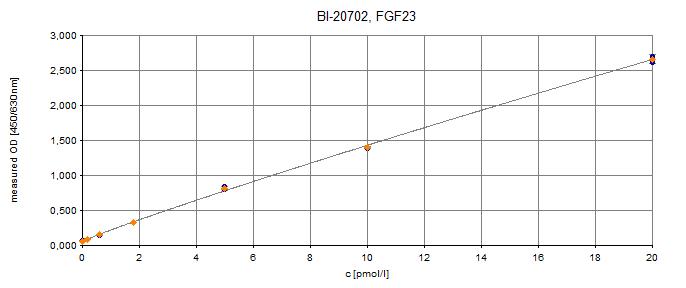
Typical Standard Curve
The figure below shows a typical standard curve for the FGF23 (C-terminal) ELISA. The immunoassay is calibrated against recombinant C-terminal FGF23 peptide:
ELISA Kit Components
|
Contents |
Description |
Quantity |
|
PLATE |
Goat polyclonal anti-FGF23 antibody pre-coated microtiter strips in strip holder, packed in aluminum bag with desiccant |
12 x 8 tests |
|
WASHBUF |
Wash buffer concentrate 20x, natural cap |
1 x 50 ml |
|
STD |
Standards 1-7, (0; 0.2; 0.6; 1.8; 5; 10; 20 pmol/l), recombinant human C-terminal FGF23 peptide in human serum, white caps, lyophilized |
7 vials |
|
CTRL |
Control A + B, yellow caps, lyophilized, exact concentrations see labels |
2 vials |
|
AB |
Rabbit polyclonal anti-FGF23 antibody – biotin labeled, green cap, green dye, ready to use |
1 x 6 ml |
|
ASYBUF |
Assay buffer, red cap, ready to use |
1 x 20 ml |
|
CONJ |
Conjugate (streptavidin-HRP), amber bottle, amber cap, ready to use |
1 x 13 ml |
|
SUB |
Substrate (TMB solution), amber bottle, blue cap, ready to use |
1 x 13 ml |
|
STOP |
Stop solution, white cap, ready to use |
1 x 7 ml |
Storage instructions: All reagents of the FGF23 (C-terminal) ELISA kit are stable at 2-8°C until the expiry date stated on the label of each reagent.
Sample Collection & Storage
Serum, EDTA plasma, heparin plasma and citrate plasma are suitable for use in this assay. Do not change sample type during studies. We recommend duplicate measurements for all samples, standards and controls. The sample collection and storage conditions listed are intended as general guidelines.
Serum & Plasma
Collect venous blood samples in standardized serum separator tubes (SST) or standardized blood collection tubes using EDTA, heparin or citrate as an anticoagulant. For serum samples, allow samples to clot for 30 minutes at room temperature. Perform separation by centrifugation according to the tube manufacturer’s instructions for use. Assay the acquired samples immediately or aliquot and store at -25°C or lower. Lipemic or haemolyzed samples may give erroneous results. Samples are stable for at least four freeze-thaw cycles.
Reagent Preparation
Wash Buffer
|
1. |
Bring the WASHBUF concentrate to room temperature. Crystals in the buffer concentrate will dissolve at room temperature. |
|
2. |
Dilute the WASHBUF concentrate 1:20, e.g. 50 ml WASHBUF + 950 ml distilled or deionized water. Only use diluted WASHBUF when performing the assay. |
The diluted WASHBUF is stable up to one month at 4°C (2-8°C).
Standards & Controls For Serum and Plasma Measurements
|
1. |
Pipette 400 µl of distilled or deionized water into each standard (STD) and control (CTRL) vial. The exact concentration is printed on the label of each vial. |
|
2. |
Leave at room temperature (18-26°C) for 15 min. Vortex gently. |
Reconstituted STDs and CTRLs are stable for 4 h at room temperature (18-24°C) or at -25°C or lower until expiry date stated on the label. STDs and CTRLs are stable for three freeze-thaw cycles.
Sample Preparation
Bring samples to room temperature and mix gently to ensure the samples are homogenous. We recommend duplicate measurements for all samples.
Samples for which the OD value exceeds the highest point of the standard range can be diluted with ASYBUF (assay buffer). If dilution is required, we recommend to dilute serum samples 1:11 (1+10, e.g. 10 µl sample + 100 µl ASYBUF) and plasma samples 1:41 (1+40, e.g. 10 µl sample + 400 µl ASYBUF). The kit includes sufficient ASYBUF for a 1:41 dilution of 40 samples. Additional ASYBUF can be ordered (cat. no. BI-201702-ASYBUF).
Assay Protocol
Read the entire protocol before beginning the assay.
|
1. |
Bring samples and reagents to room temperature (18-24°C). |
|
2. |
Mark positions for STD/CTRL/SAMPLE (standard/control/sample) on the protocol sheet. |
|
3. |
Take microtiter strips out of the aluminum bag. Store unused strips with desiccant at 4°C in the aluminum bag. Strips are stable until expiry date stated on the label. |
|
4. |
Pipette 50 µl STD/SAMPLE/CTRL (Standard/Sample/Control) in duplicate into respective well. |
|
5. |
Add 50 µl AB (biotinylated anti-FGF23 antibody, green cap, green dye) into each well, mix gently. |
|
6. |
Cover the plate tightly and incubate overnight (20-24 h) at room temperature (18-24°C). |
|
7. |
Aspirate and wash wells 5x with 300 µl diluted WASHBUF. After the final wash, remove remaining WASHBUF by strongly tapping the plate against a paper towel. |
|
8. |
Add 100 µl CONJ (conjugate, amber cap) into each well. |
|
9. |
Cover the plate tightly and incubate for 1 h at room temperature (18-24°C) in the dark. |
|
10. |
Aspirate and wash wells 5x with 300 µl diluted WASHBUF. After the final wash, remove remaining WASHBUF by strongly tapping the plate against a paper towel. |
|
11. |
Add 100 µl SUB (substrate, blue cap) into each well. |
|
12. |
Incubate for 30 min at room temperature (18-24°C) in the dark. |
|
13. |
Add 50 µl STOP (stop solution, white cap) into each well, mix gently. |
|
14. |
Measure absorbance immediately at 450 nm with reference 630 nm, if available. |
Calculation of Results
Construct a standard curve from the absorbance read-outs of the standards using commercially available software capable of generating a four-parameter logistic (4-PL) fit. Alternatively, plot the standards’ concentration on the x-axis against the mean absorbance for each standard on the y-axis and draw a best fit curve through the points on the graph. Curve fitting algorithms other than 4-PL have not been validated and will need to be evaluated by the user.
Obtain sample concentrations from the standard curve. If required, pmol/l can be converted into pg/ml by applying a conversion factor (1 pg/ml = 0.133 pmol/l; MW: 7.52 kDa). Respective dilution factors have to be considered when calculating the final concentration of the sample.
The quality control protocol supplied with the kit shows the results of the final release QC for each kit lot. Data for optical density obtained by customers may differ due to various influences and/or due to the normal decrease of signal intensity during shelf life. However, this does not affect validity of results as long as an OD of 1.50 or more is obtained for the standard with the highest concentration and the values of the CTRLs are in range (target ranges see labels).
INFORMATION ON THE ANALYTE
FGF23 protein
FGF23 (fibroblast growth factor 23) is a member of the fibroblast growth factor family and controls phosphate and vitamin D homeostasis. It is a 32 kDa protein of 251 amino acids including a 24 amino acid signal peptide. The N-terminal FGF homology region of FGF23 is separated from the unique C-terminal region by a proteolytic cleavage site. A proportion of FG23 is proteolytically processed between arginine179 and serine180 to generate N-terminal and C-terminal fragments. Therefore, the major forms of FGF23 present in human circulation are hormonally intact FGF23 and inactive N-terminal and C-terminal fragments. FGF23 binds to FGF receptor 1c (FGFR1c) with its N-terminal region, while the C-terminal region is capable of interacting with the co-receptor αKlotho to confer high-affinity binding to the receptor. FGFR1c and αKlotho are expressed in the distal nephron and the parathyroid gland. Co-receptor independent signaling of FGF23 has been described for other FGFRs, which are expressed in a variety of tissues. The main source of FGF23 are osteocytes in the bone.
|
Molecular Weight |
7.52 kDa (C-terminal fragment of FGF23) |
|
Cellular localization |
Extracellular |
|
Post-translational modifications |
Glycosylation, phosphorylation |
|
Sequence similarities |
Member of the fibroblast growth factor (FGF) family |
|
Alternative Names |
Fibroblast growth factor 23, ADHR, FGFN, HPDR2, HYPF, PHPTC, HFTC2 |
|
Entrez/NCBI ID |
8047 link: https://www.ncbi.nlm.nih.gov/gene/8074 |
|
Genecards |
GC12M004347 link: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGF23 |
|
OMIM |
605380 link: https://www.omim.org/entry/605380 |
|
PDB |
2P39 link: https://www.rcsb.org/structure/2P39 |
|
Pfam |
PF00167 link: https://pfam.xfam.org/family/PF00167 |
|
Protein Atlas |
FGF23 link: https://www.proteinatlas.org/ENSG00000118972-FGF23/tissue |
|
Uniprot ID |
Q9GZV9 link: https://www.uniprot.org/uniprot/Q9GZV9 |
FGF23 Function
FGF23 is an endocrine hormone regulating phosphate homeostasis by modulating renal phosphate reabsorption, vitamin D metabolism and parathyroid hormone (PTH) secretion. Increased dietary phosphate intake or reduced phosphate excretion due to renal insufficiency stimulates FGF23 secretion in the bone. FGF23 induces the internalization of phosphate transporters in the kidney and thus, reduces phosphate reabsorption. The subsequent renal excretion of phosphate (phosphaturia) restores phosphate balance. PTH and active vitamin D (1,25-dihydroxyvitamin D) both stimulate FGF23 secretion and are suppressed by rising FGF23 levels in a negative feedback loop. FGF23 inhibits the activation of 25-hydroxyvitamin D into 1,25 dihydroxyvitamin D through downregulation of the enzyme 1α-hydroxylase catalyzing this metabolic step. At the same time, FGF23 promotes the upregulation of 24-hydroxylase, an enzyme mediating the inactivation of 1,25 dihydroxyvitamin D. FGF23-induced downregulation of active vitamin D limits the gastrointestinal uptake of phosphate and thus, contributes to the phosphate lowering effects of FGF23. Since PTH releases phosphate from bone, FGF23-mediated suppression of PTH secretion builds the third axis by which phosphate homeostasis is controlled by FGF23.
In humans, circulating levels of FGF23 can be altered in both genetic and acquired diseases. A primary excess in circulating intact FGF23 is the underlying cause of diseases like X-linked hypophosphatemic rickets (XLH), autosomal dominant/recessive hypophosphatemic rickets (ADHR/ARHR) or tumor-induced osteomalacia. The excessive FGF23 levels in these diseases cause renal phosphate wasting, low active vitamin D concentrations and defective mineralization of bones. In case of a genetic disease, the mutations accounting for hyperphosphatemia or hypophosphatemia are located in either the FGF23 gene itself or one of its regulators. In ADHR, missense mutations in the FGF23 gene inhibit the cleavage into N- and C-terminal fragments and thus, increase the amount of circulating intact FGF23. However, the exact molecular mechanisms by which some mutations other than in the FGF23 gene itself are causing FGF23-related disease is not yet fully understood.
Circulating FGF23 is also elevated in patients with chronic kidney disease (CKD), presumably in response to decreased renal excretion of phosphate. During the course of the disease, FGF23 secretion is upregulated and the ratio between C-terminal and intact FGF23 is shifted towards the intact protein. Simultaneous upregulation of FGF23 cleavage in osteocytes maintains near-normal levels of biologically active, intact circulating FGF23, whereas downregulated or impaired FGF23 cleavage may contribute to elevated intact serum FGF23 in CKD. The increase in FGF23 concentration in CKD patients is accompanied by low levels of active vitamin D and secondary hyperparathyroidism. Increased FGF23 in CKD patients is not only associated with disease progression, but also with left ventricular hypertrophy and endothelial dysfunction.
Besides hyperphophatemia, other conditions associated with CKD such as acidosis, tissue hypoxia, iron deficiency, and inflammation may also contribute to the increase in FGF23 levels. Iron stimulates FGF23 production and is counterbalanced by increased cleavage of newly synthesized FGF23 within healthy osteocytes. As a result, FGF23 fragments are elevated, but serum phosphate levels remain normal due to normal circulating levels of biologically active FGF23. Inflammation plays an essential role in FGF23 regulation as it triggers FGF23 expression and cleavage thus revealing its pro-inflammatory and immune-modulatory properties.
Epidemiological data suggests that higher FGF23 concentrations are associated with all-cause mortality, cardiovascular mortality, a higher risk of myocardial infarction, stroke and heart failure. Furthermore, FGF23 levels could induce left ventricular hypertrophy, potentially via direct but FGFR1c- and α-klotho-independent signaling in the heart.
-
Bone Diseases
-
Autosomal dominant hypophosphatemic rickets (ADHR)
-
Autosomal recessive hypophosphatemic rickets (ARHR)
-
X-linked hypophosphatemic rickets (XLH)
-
Autosomal recessive hypercholesterolaemia rickets (ARHR)
-
McCune-Albright syndrome/fibrous dysplasia
-
Hypophosphatemic rickets and hyperparathyroidism (HRHPT)
-
Drug-induced rickets/osteomalacia
-
Hyperphosphatemia
-
Hyperostosis-hyperphosphatemia syndrome
-
Fibrous dysplasia
-
Neurofibromatosis
-
Osteoglophonic dysplasia (OGD)
-
Linear nevus sebaceous syndrome (LNSS)/epidermal nevus syndrome (ENS)
-
Jansen-type metaphyseal chondrocdysplasia
-
-
Cancer
-
Tumor-induced osteomalacia
-
Familial tumor calcinosis
-
-
Cardiovascular disease
-
Heart failure
-
Left-ventricular hypertrophy
-
Vascular calcification
-
-
Endocrine disorders
-
Primary hyperparathyroidism
-
Secondary hyperparathyroidism
-
-
Kidney Disease
-
Chronic kidney disease (CKD)
-
Chronic kidney disease-mineral and bone disorder (CKD-MBD)
-
Chronic kidney disease and cardiovascular disease
-
Acute kidney injury
-
-
Metabolic disorders
-
Dyslipidemia
-
Insulin resistance/ type 2 diabetes
-
Anemia
-
-
α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling.
Chen, G., Liu, Y., Goetz, R., Fu, L., Jayaraman, S., Hu, M.-C., Moe, O.W., Liang, G., Li, X., Mohammadi, M., 2018. Nature 553, 461–466.
PMID:29342138
-
Physiology of FGF23 and overview of genetic diseases associated with renal phosphate wasting
Bachhetta, J., Bardet, C., Prié, D., 2019. Metab. Clin. Exp.
PMCID: PMC6007875
PMID:30664852
-
FGF23 beyond Phosphotropic Hormone
Takashi, Y., Fukumoto, S., 2018. Trends Endocrinol. Metab.
PMID:30217676
-
FGF23 in Cardiovascular Disease: Innocent Bystander or Active Mediator?
Stöhr, R., Schuh, A., Heine, G.H., Brandenburg, V., 2018. Front. Endocrinol.
PMCID: PMC6036253
PMID:30013515
-
FGF23 Actions on Target Tissues—With and Without Klotho
Richter, B., Faul, C., 2018. Front. Endocrinol.
PMCID: PMC5940753
PMID:29770125
-
Physiological Actions of Fibroblast Growth Factor-23
Erben, R., 2018. Front. Endocrinol.
PMCID: PMC5985418
PMID:29892265
All Biomedica ELISAs are validated according to FDA/ICH/EMEA guidelines. For more information about our validation guidelines, please refer to our quality page and published validation guidelines and literature.
-
ICH Q2(R1) Validation of Analytical Procedures: Text and Methodology.
-
EMEA/CHMP/EWP/192217/2009 Guideline on bioanalytical method validation.
-
Bioanalytical Method Validation, Guidance for Industry, FDA, May 2018
Calibration
The FGF23 (C-terminal) immunoassay is calibrated against C-terminal FGF23 fragment (AA 180-251 of FGF23 (https://www.uniprot.org/uniprot/Q9GZV9)).
Detection Limit & Sensitivity
To determine the sensitivity of the FGF23 (C-terminal) ELISA, experiments measuring the lower limit of detection (LOD) and the lower limit of quantification (LLOQ) were conducted.
The LOD, also called the detection limit, is the lowest point at which a signal can be distinguished above the background signal, i.e. the signal that is measured in the absence of FGF23, with a confidence level of 99%. It is defined as the mean back calculated concentration of standard 1 (0 pmol/l of FGF23, five independent measurements) plus three times the standard deviation of the measurements.
The LLOQ, or sensitivity of an assay, is the lowest concentration at which an analyte can be accurately quantified. The criteria for accurate quantification at the LLOQ are an analyte recovery between 75 and 125% and a coefficient of variation (CV) of less than 25%. To determine the LLOQ, standard 2, i.e. the lowest standards containing FGF23, is diluted, measured five times and its concentration back calculated. The lowest dilution, which meets both criteria, is reported as the LLOQ.
The following values were determined for the FGF23 (C-terminal) ELISA:
|
LOD |
0.08 pmol/l |
|
LLOQ |
0.1 pmol/l |
Precision
The precision of an ELISA is defined as its ability to measure the same concentration consistently within the same experiments carried out by one operator (within-run precision or repeatability) and across several experiments using the same samples but conducted by several operators at different locations using different ELISA lots (in-between-run precision or reproducibility).
Within-Run Precision
Within-run precision was tested by measuring two samples six times within one FGF23 (C-terminal) ELISA lot. The experiment was conducted by one operator.
|
ID |
n |
Mean FGF23 [pmol/l] |
SD [pmol/l] |
CV [%] |
|
Sample 1 |
6 |
0.6 |
0.07 |
12 |
|
Sample 2 |
6 |
10.0 |
0.6 |
6 |
In-Between-Run Precision
In-between-run precision was assessed by measuring two samples ten times within two different FGF23 (C-terminal) ELISA kit lots by four different operators.
|
n |
Mean FGF23 [pmol/l] |
SD [pmol/l] |
CV [%] |
|
|
Sample 1 |
10 |
0.61 |
0.06 |
10 |
|
Sample 2 |
10 |
9.91 |
0.5 |
5 |
Accuracy
The accuracy of an ELISA is defined as the precision with which it can recover samples of known concentrations.
The recovery of the FGF23 (C-terminal) ELISA was measured by adding recombinant C-terminal FGF23 fragment to samples containing a known concentration endogenous FGF23. The %recovery of the spiked concentration was calculated as the percentage of measured compared over the expected value.
This table shows the summary of the recovery experiments in the FGF23 (C-terminal) ELISA in different sample matrices:
|
% Recovery |
|||||
|
Sample Matrix |
n |
+5 pmol/l |
+10 pmol/l |
||
|
Mean |
Range |
Mean |
Range |
||
|
Serum |
13 |
96 |
84-108 |
89 |
60-103 |
|
EDTA plasma |
7 |
97 |
73-123 |
94 |
81-114 |
|
Citrate plasma |
7 |
100 |
61-132 |
90 |
58-107 |
|
Heparin plasma |
8 |
101 |
68-161 |
92 |
69-118 |
Data showing recovery (R [%]) of recombinant C-terminal FGF23 fragment in human serum samples:
|
FGF23 [pmol/l] |
% Recovery |
|||||||
|
Sample Matrix |
ID |
Reference |
+5 pmol/l |
+10 pmol/l |
+5 pmol/l |
+10 pmol/l |
||
|
Serum |
s1 |
1.3 |
6.4 |
10.1 |
103 |
87 |
||
|
Serum |
s2 |
0.8 |
5.9 |
11.0 |
102 |
101 |
||
|
Serum |
s3 |
2.2 |
7.0 |
10.4 |
96 |
82 |
||
|
Serum |
s4 |
4.8 |
10.2 |
13.9 |
108 |
91 |
||
|
Serum |
s5 |
2.1 |
7.1 |
12.4 |
100 |
103 |
||
|
Serum |
s6 |
1.8 |
6.1 |
10.6 |
85 |
88 |
||
|
Serum |
s7 |
1.2 |
5.5 |
7.3 |
84 |
60 |
||
|
Serum |
s8 |
1.6 |
6.2 |
11.4 |
92 |
98 |
||
|
Serum |
s9 |
2.7 |
7.2 |
11.7 |
90 |
90 |
||
|
Serum |
s10 |
0.8 |
5.4 |
10.1 |
92 |
92 |
||
|
Serum |
s11 |
1.1 |
5.9 |
10.4 |
95 |
93 |
||
|
Serum |
s12 |
0.6 |
5.6 |
9.3 |
101 |
87 |
||
|
Serum |
s13 |
1.9 |
7.1 |
10.4 |
104 |
85 |
||
|
96 |
89 |
Mean |
||||||
|
84 |
60 |
Min |
||||||
|
108 |
103 |
Max |
||||||
Data showing recovery of recombinant C-terminal FGF23 fragment in human EDTA plasma samples:
|
FGF23 [pmol/l] |
Recovery [%] |
|||||||
|
Sample Matrix |
ID |
Reference |
+5 pmol/l |
+10 pmol/l |
+5 pmol/l |
+10 pmol/l |
||
|
EDTA plasma |
e1 |
1.3 |
6.7 |
11.9 |
108 |
106 |
||
|
EDTA plasma |
e2 |
1.4 |
5.0 |
9.5 |
73 |
81 |
||
|
EDTA plasma |
e3 |
1.8 |
6.8 |
13.2 |
101 |
114 |
||
|
EDTA plasma |
e4 |
1.2 |
5.4 |
8.9 |
84 |
77 |
||
|
EDTA plasma |
e5 |
3.2 |
7.6 |
11.3 |
88 |
81 |
||
|
EDTA plasma |
e6 |
2.0 |
7.0 |
11.6 |
100 |
96 |
||
|
EDTA plasma |
e7 |
1.3 |
7.4 |
11.3 |
123 |
101 |
||
|
97 |
94 |
Mean |
||||||
|
73 |
77 |
Min |
||||||
|
123 |
114 |
Max |
||||||
Data showing recovery of recombinant C-terminal FGF23 fragment in human citrate plasma samples:
|
FGF23 [pmol/l] |
Recovery [%] |
|||||||
|
Sample Matrix |
ID |
Reference |
+5 pmol/l |
+10 pmol/l |
+5 pmol/l |
+10 pmol/l |
||
|
Citrate plasma |
c1 |
4.7 |
10.5 |
14.9 |
116 |
101 |
||
|
Citrate plasma |
c2 |
2.2 |
8.8 |
12.0 |
132 |
98 |
||
|
Citrate plasma |
c3 |
1.8 |
8.0 |
12.1 |
124 |
103 |
||
|
Citrate plasma |
c4 |
1.8 |
7.2 |
12.5 |
110 |
107 |
||
|
Citrate plasma |
c5 |
1.8 |
5.4 |
9.2 |
74 |
74 |
||
|
Citrate plasma |
c6 |
2.7 |
5.8 |
8.6 |
61 |
58 |
||
|
Citrate plasma |
c7 |
3.9 |
8.0 |
12.7 |
82 |
88 |
||
|
100 |
90 |
Mean |
||||||
|
61 |
58 |
Min |
||||||
|
132 |
107 |
Max |
||||||
Data showing recovery of recombinant C-terminal FGF23 fragment in human heparin plasma samples:
|
FGF23 [pmol/l] |
Recovery [%] |
|||||||
|
Sample Matrix |
ID |
Reference |
+5 pmol/l |
+10 pmol/l |
+5 pmol/l |
+10 pmol/l |
||
|
Heparin plasma |
c1 |
1.8 |
9.9 |
13.7 |
161 |
118 |
||
|
Heparin plasma |
c2 |
2.2 |
7.1 |
11.5 |
97 |
93 |
||
|
Heparin plasma |
c3 |
1.3 |
5.5 |
9.1 |
84 |
78 |
||
|
Heparin plasma |
c4 |
2.2 |
7.1 |
11.7 |
98 |
95 |
||
|
Heparin plasma |
c5 |
2.2 |
6.3 |
10.2 |
82 |
80 |
||
|
Heparin plasma |
c6 |
1.2 |
4.6 |
8.1 |
68 |
69 |
||
|
Heparin plasma |
c7 |
1.8 |
7.3 |
12.3 |
110 |
105 |
||
|
Heparin plasma |
c8 |
2.2 |
7.5 |
12.1 |
106 |
99 |
||
|
101 |
92 |
Mean |
||||||
|
68 |
78 |
Min |
||||||
|
161 |
118 |
Max |
||||||
Dilutuion Linearity & Paralellism
Tests of dilution linearity and parallelism ensure that both endogenous and recombinant samples containing FGF23 behave in a dose dependent manner and are not affected by matrix effects. Dilution linearity assesses the accuracy of measurements in diluted human samples spiked with known concentrations of recombinant analyte. By contrast, parallelism refers to dilution linearity in human samples and provides evidence that the endogenous analyte behaves in the same way as the recombinant one. Dilution linearity and parallelism are assessed for each sample type and should be within 20% of the expected concentration.
Parallelism was assessed by serially diluting clinical samples containing endogenous FGF23 with assay buffer.
The table below shows the mean recovery and range of serially diluted endogenous FGF23 in several sample matrices:
|
% Recovery of endogenous FGF23 in diluted samples |
|||||||
|
Sample Matrix |
n |
1+1 |
1+3 |
1+7 |
|||
|
Mean |
Range |
Mean |
Range |
Mean |
Range |
||
|
Serum |
9 |
105 |
93-113 |
100 |
89-126 |
108 |
91-124 |
|
EDTA plasma |
4 |
103 |
67-127 |
103 |
69-120 |
106 |
68-124 |
|
Citrate plasma |
5 |
102 |
94-109 |
106 |
93-123 |
101 |
93-118 |
|
Heparin plasma |
10 |
102 |
92-108 |
106 |
93-117 |
104 |
90-132 |
Data showing dilution linearity of endogenous FGF23 in human serum samples:
|
FGF23 [pmol/l] |
% Recovery |
|||||||||
|
Sample matrix |
ID |
Reference |
1+1 |
1+3 |
1+7 |
1+1 |
1+3 |
1+7 |
||
|
Serum |
s1 |
3.8 |
2.1 |
1.1 |
0.6 |
111 |
117 |
124 |
||
|
Serum |
s2 |
16.2 |
8.1 |
3.6 |
1.9 |
99 |
89 |
92 |
||
|
Serum |
s3 |
7.0 |
3.5 |
1.8 |
0.9 |
100 |
103 |
101 |
||
|
Serum |
s4 |
9.0 |
4.6 |
2.1 |
1.0 |
102 |
95 |
91 |
||
|
Serum |
s5 |
8.5 |
4.7 |
2.7 |
1.3 |
111 |
126 |
123 |
||
|
Serum |
s6 |
6.0 |
3.4 |
1.3 |
0.9 |
113 |
90 |
121 |
||
|
Serum |
s7 |
15.7 |
7.3 |
3.5 |
1.9 |
93 |
89 |
96 |
||
|
Serum |
s8 |
20.4 |
10.9 |
5.1 |
2.6 |
106 |
101 |
103 |
||
|
Serum |
s9 |
10.6 |
5.6 |
2.5 |
1.6 |
105 |
93 |
121 |
||
|
105 |
100 |
108 |
Mean |
|||||||
|
93 |
89 |
91 |
Min |
|||||||
|
113 |
126 |
124 |
Max |
|||||||
Data showing dilution linearity of endogenous FGF23 in human EDTA plasma samples:
|
FGF23 [pmol/l] |
% Recovery |
||||||||
|
Sample Matrix |
ID |
Reference |
1+1 |
1+3 |
1+7 |
1+1 |
1+3 |
1+7 |
|
|
EDTA plasma |
e1 |
3.8 |
2.4 |
1.1 |
0.6 |
127 |
120 |
121 |
|
|
EDTA plasma |
e2 |
23.3 |
13.6 |
6.7 |
3.2 |
117 |
115 |
112 |
|
|
EDTA plasma |
e3 |
8.9 |
4.5 |
2.4 |
1.4 |
101 |
109 |
124 |
|
|
EDTA plasma |
e4 |
20.7 |
7.0 |
3.6 |
1.8 |
67 |
69 |
68 |
|
|
103 |
103 |
106 |
Mean |
||||||
|
67 |
69 |
68 |
Min |
||||||
|
127 |
120 |
124 |
Max |
||||||
Data showing recovery of endogenous FGF23 in a human citrate plasma sample:
|
FGF23 [pmol/l] |
% Recovery |
||||||||
|
Sample Matrix |
ID |
Reference |
1+1 |
1+3 |
1+7 |
1+1 |
1+3 |
1+7 |
|
|
Citrate plasma |
e1 |
17.0 |
8.0 |
4.0 |
2.0 |
94 |
93 |
93 |
|
|
Citrate plasma |
e2 |
13.0 |
7.1 |
4.0 |
1.9 |
109 |
123 |
118 |
|
|
Citrate plasma |
e3 |
12.6 |
6.3 |
3.1 |
1.6 |
100 |
100 |
101 |
|
|
Citrate plasma |
e4 |
10.0 |
5.3 |
2.6 |
1.2 |
106 |
106 |
97 |
|
|
Citrate plasma |
e5 |
9.7 |
4.8 |
2.6 |
1.2 |
100 |
108 |
97 |
|
|
102 |
106 |
101 |
Mean |
||||||
|
94 |
93 |
93 |
Min |
||||||
|
109 |
123 |
118 |
Max |
||||||
Data showing recovery of endogenous FGF23 in a human heparin plasma sample:
|
FGF23 [pmol/l] |
% Recovery |
||||||||
|
Sample matrix |
ID |
Reference |
1+1 |
1+3 |
1+7 |
1+1 |
1+3 |
1+7 |
|
|
Heparin plasma |
s1 |
7.5 |
4.1 |
2.2 |
1.1 |
108 |
117 |
119 |
|
|
Heparin plasma |
s2 |
5.8 |
2.9 |
1.7 |
1.0 |
102 |
117 |
132 |
|
|
Heparin plasma |
s3 |
11.4 |
6.4 |
3.3 |
1.6 |
113 |
117 |
112 |
|
|
Heparin plasma |
s4 |
9.0 |
4.9 |
2.6 |
1.3 |
113 |
117 |
112 |
|
|
Heparin plasma |
s5 |
7.7 |
3.6 |
1.8 |
0.9 |
92 |
93 |
90 |
|
|
Heparin plasma |
s6 |
10.7 |
5.3 |
2.6 |
1.2 |
100 |
97 |
91 |
|
|
Heparin plasma |
s7 |
7.6 |
3.9 |
2.1 |
1.0 |
102 |
111 |
108 |
|
|
Heparin plasma |
s8 |
6.4 |
3.2 |
1.5 |
0.8 |
99 |
93 |
95 |
|
|
Heparin plasma |
s9 |
3.7 |
1.8 |
1.0 |
0.4 |
98 |
109 |
93 |
|
|
Heparin plasma |
s10 |
8.6 |
4.1 |
2.0 |
1.0 |
96 |
95 |
90 |
|
|
102 |
106 |
104 |
Mean |
||||||
|
92 |
93 |
91 |
Min |
||||||
|
113 |
117 |
132 |
Max |
||||||
Specificity
The specificity of an ELISA is defined as its ability to exclusively recognize the analyte of interest. The specificity of the FGF23 (C-terminal) ELISA was shown by characterizing both the capture and the detection antibodies through affinity measurements. In addition, the specificity of the ELISA was established through competition experiments, which measure the ability of the antibodies to exclusively bind to FGF23.
Isoforms
There are no isoforms of FGF23 known.
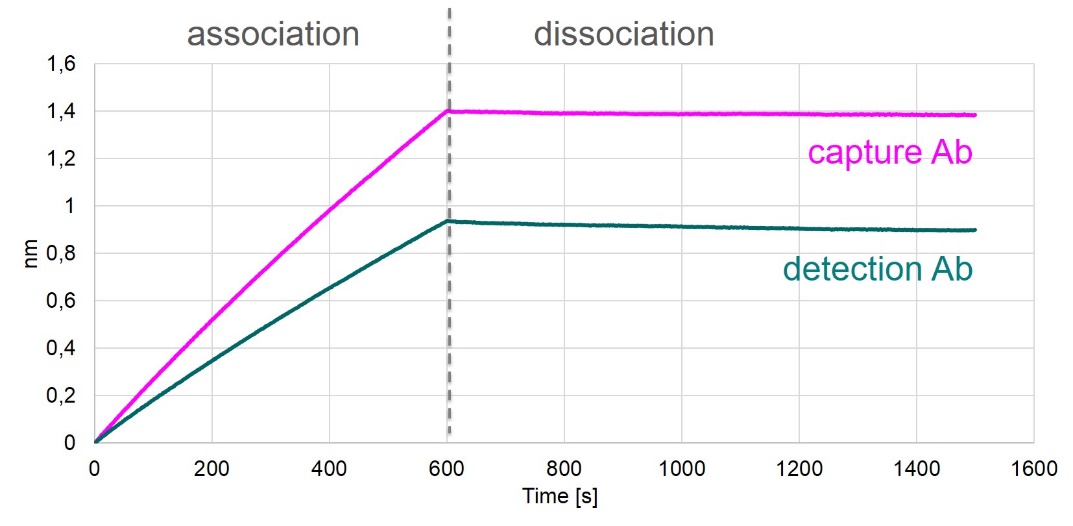
Affinities of Coating and Detection Antibodies
Antibody affinities to FGF23 were tested by biolayer interferometry measurements (Octet), which measures the binding of antibodies to a FGF23-coated sensor.
The results of these measurements are shown in the figure below.
Both antibodies used in the FGF23 (C-terminal) ELISA bind to FGF23 with high affinity.
Competition of Signal
Competition experiments were carried out by pre-incubating human samples with an excess of capture antibody. The concentration measured in this mixture was then compared to a reference value, which was obtained from the same sample but without the pre-incubation step. Mean competition was 95%.
|
FGF23 [pmol/l] |
% Competition |
||||
|
Sample matrix |
ID |
Reference |
Reference + capture AB |
||
|
Serum |
s1 |
2,7 |
0 |
100 |
|
|
Serum |
s2 |
0,8 |
0,2 |
77 |
|
|
Serum |
s3 |
5,6 |
0,1 |
98 |
|
|
Serum |
s4 |
0,8 |
0 |
100 |
|
|
Serum |
s5 |
0,5 |
0 |
100 |
|
|
Serum |
s6 |
0,3 |
0 |
100 |
|
|
Serum |
s7 |
0,3 |
0 |
100 |
|
|
EDTA plasma |
e1 |
11,5 |
0,7 |
93 |
|
|
EDTA plasma |
e2 |
8,5 |
0,5 |
94 |
|
|
EDTA plasma |
e3 |
17,2 |
0,3 |
99 |
|
|
EDTA plasma |
e4 |
8,9 |
0,2 |
98 |
|
|
Heparin Plasma |
h1 |
10,5 |
0,4 |
96 |
|
|
Heparin plasma |
h2 |
6,0 |
0,4 |
93 |
|
|
Heparin plasma |
h3 |
5,3 |
0,2 |
96 |
|
|
Heparin plasma |
h4 |
3,1 |
0,4 |
88 |
|
|
95 |
Mean |
Sample Stability
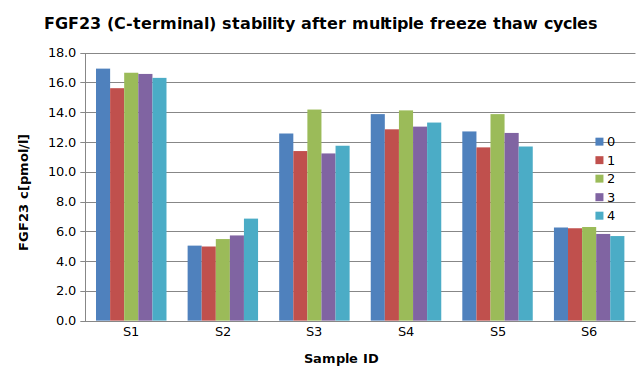
The stability of endogenous C-terminal FGF23 fragment was tested by comparing five measurements in samples that had undergone four freeze-thaw cycles.
For freeze-thaw experiments, samples were collected according to the supplier’s instruction using blood collection devices and stored at -80°C. Reference samples were freeze-thawed once. The mean recovery of sample concentration after four freeze-thaw cycles is 101%.
|
FGF23 [pmol/l] |
% Recovery after 4 freeze/thaw cycles | |||||||
|
Sample matrix |
ID |
Reference |
1x |
2x |
3x |
4x |
||
|
Serum |
s1 |
16,9 |
15,6 |
16,7 |
16,6 |
16,3 |
96% |
|
|
Serum |
s2 |
5,0 |
5,0 |
5,5 |
5,7 |
6,8 |
136% |
|
|
Serum |
s3 |
12,6 |
11,4 |
14,2 |
11,2 |
11,7 |
93% |
|
|
Serum |
s4 |
13,9 |
12,9 |
14,1 |
13,0 |
13,3 |
96% |
|
|
Serum |
s5 |
12,7 |
11,6 |
13,9 |
12,6 |
11,7 |
92% |
|
|
Serum |
s6 |
6,3 |
6,2 |
6,3 |
5,8 |
5,7 |
91% |
|
|
101 |
Mean |
|||||||
All samples should undergo a maximum of four freeze-thaw cycles.
Sample Values
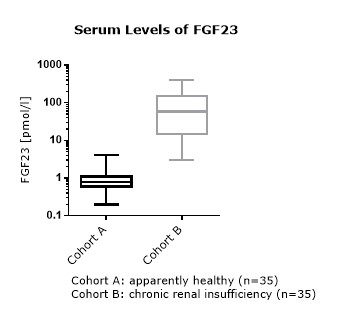
FGF23 Values in Apparently Healthy Individuals
To provide expected values for circulating FGF23, a panel of samples from apparently healthy donors was tested.
A summary of the results is shown below:
|
FGF23 (C-terminal) [pmol/l] |
|||||||
|
Sample Matrix |
n |
Mean |
Median |
5% Percentile |
95% Percentile |
Minimum |
Maximum |
|
Serum |
35 |
1.1 |
0.8* |
0.3 |
3.0 |
0.2 |
4.2 |
|
EDTA plasma |
22 |
1.6 |
1.3 |
0.6 |
4.0 |
0.3 |
4.8 |
|
Heparin plasma |
22 |
1.5 |
1.2 |
0.5 |
3.8 |
0.3 |
4.0 |
|
Citrate plasma |
30 |
1.9 |
1.4** |
0.9 |
4.5 |
0.7 |
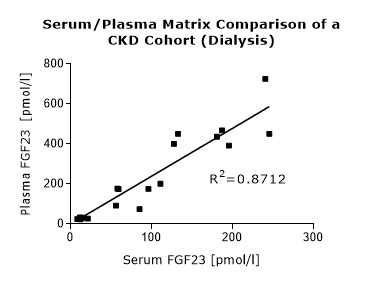
6.8 |
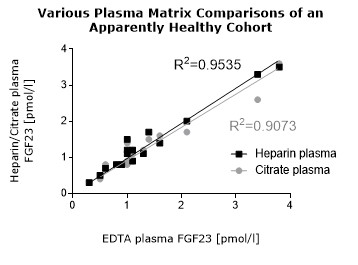
* Serum and plasma values of FGF23 (C-terminal) show a correlation of R2= 0.8675 (see matrix comparison data).
**Various plasma matrices (EDTA, heparin, citrate) show a correlation of R2= 0.9535 (see matrix comparison data).
It is recommended to establish the normal range for each laboratory.
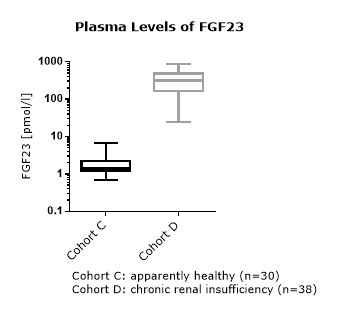
FGF23 (C-terminal) Values in Chronic Kidney Disease and Dialysis Panels
In addition to samples from apparently healthy donors, several panels of samples from chronic kidney disease (CKD) and dialysis patients were tested.
Summary of the results obtained with several chronic kidney disease (dialysis) panels:
|
FGF23 (C-terminal) [pmol/l] |
||||||||
|
Sample Matrix |
n |
Panel |
Mean |
Median |
5% Percentile |
95% Percentile |
Minimum |
Maximum |
|
Serum |
18 |
CKD |
8.2 |
5.7 |
1.4 |
20 |
1.4 |
20.0 |
|
Serum |
20 |
Dialysis |
89 |
59 |
6 |
307 |
6 |
310 |
|
Serum |
15 |
Dialysis |
96 |
20 |
3 |
408 |
3 |
408 |
|
Plasma |
20 |
Dialysis |
248 |
177 |
24 |
717 |
24 |
725 |
|
Heparin plasma |
18 |
Dialysis |
441 |
355 |
150 |
882 |
150 |
882 |
Comparison of FGF23 (C-terminal) Values in Apparently Healthy Individuals and Patients with Chronic Kidney Disease (CKD)/ Dialysis
Matrix Comparison
To assess whether all tested matrices behave the same way in the FGF23 (C-terminal) ELISA, concentrations of FGF23 were measured in serum, EDTA, heparin and citrate plasma samples prepared from 22 apparently healthy donors. Each individual donated blood in all tested sample matrices. In addition, FGF23 concentrations were assessed in serum and plasma and plasma samples prepared from 20 patients with chronic kidney disease (dialysis). Correlations between serum and plasma samples were calculated for each cohort.
Correlations of FGF23 measurements in serum and plasma samples is shown below:
Both serum and plasma can be measured with the FGF23 (C-terminal) ELISA.
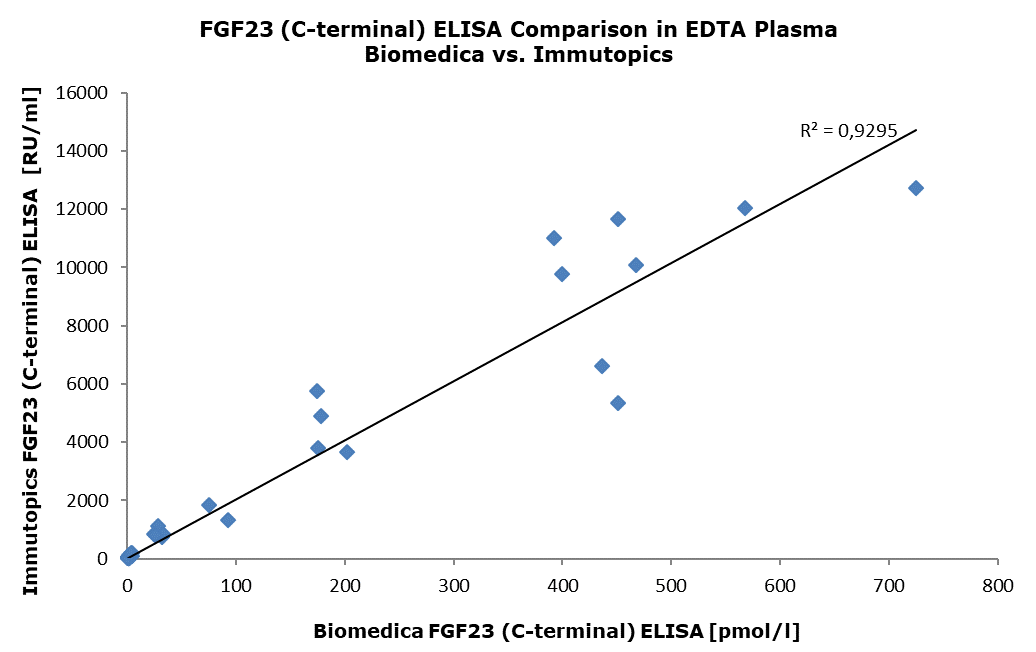
Comparison With Other FGF23 (C-terminal) ELISA Assays
The FGF23 (C-terminal) ELISA (cat. no. BI-20702) was compared with the FGF23 C-Term ELISA (cat. no. 60-6100) manufactured by Immutopics. The same panel of samples, consisting of EDTA plasma samples from apparently healthy donors (n=33) and patients with chronic kidney disease (dialysis) (n=20), was tested. The correlation between the two assays was R2=0.9295.
The figure below shows the measured concentrations in both tests:
-
Fibroblast growth factor 23 and lipid metabolism association in chronic kidney disease.
Chaikovska, M.I., Martynyuk, L.P., 2018. 1 34–40.
-
Lyngbakken, M.N., Pervez, M.O., Brynildsen, J., Pedersen, M.H., Sølvernes, J., Christensen, G., Høiseth, A.D., Omland, T., Røsjø, H., 2018. Clin. Biochem. 52, 41–47.
PMID:29074091
-
Nakayama, M., Tani, Y., Zhu, W.-J., Watanabe, K., Yokoyama, K., Fukagawa, M., Akiba, T., Wolf, M., Hirakata, H., 2018. Kidney Int Rep 3, 364–373.
PMCID: PMC5932126
PMID:29725640
-
Left Ventricular Structure in Patients With Mild-to-Moderate CKD—a Magnetic Resonance Imaging Study.
Schneider, M.P., Scheppach, J.B., Raff, U., Toncar, S., Ritter, C., Klink, T., Störk, S., Wanner, C., Schlieper, G., Saritas, T., Reinartz, S.D., Floege, J., Friedrich, N., Janka, R., Uder, M., Schmieder, R.E., Eckardt, K.-U., 2018. Kidney International Reports.
-
Effects of vitamin D supplementation on FGF23: a randomized-controlled trial.
Trummer, C., Schwetz, V., Pandis, M., Grübler, M.R., Verheyen, N., Gaksch, M., Zittermann, A., März, W., Aberer, F., Steinkellner, J., Friedl, C., Brandenburg, V., Voelkl, J., Alesutan, I., Obermayer-Pietsch, B., Pieber, T.R., Tomaschitz, A., Pilz, S., 2018. Eur J Nutr.
PMID:29602956
-
Sex and Iron Modify Fibroblast Growth Factor 23 Concentration in 1-Year-Old Children.
Holmlund-Suila, E., Enlund-Cerullo, M., Valkama, S., Hauta-Alus, H., Rosendahl, J., Helve, O., Hytinantti, T., Viljakainen, H., Andersson, S., Mäkitie, O., 2017. J. Clin. Endocrinol. Metab. 102, 4526–4533.
PMID:29029193
-
Mettang, T., Kunzmann, K., Roth, H.-J., Weisshaar, E., 2017. Acta Derm. Venereol. 97, 381–382.
PMID:27671605
-
Long-Term Effects of Severe Burn Injury on Bone Turnover and Microarchitecture.
Muschitz, G.K., Schwabegger, E., Fochtmann, A., Baierl, A., Kocijan, R., Haschka, J., Gruther, W., Schanda, J.E., Resch, H., Rath, T., Pietschmann, P., Muschitz, C., 2017. Journal of Bone and Mineral Research 32, 2381–2393.
-
Yamashita, K., Mizuiri, S., Nishizawa, Y., Kenichiro, S., Doi, S., Masaki, T., 2017. Nephrology (Carlton) 22, 947–953.
PMCID: PMC5725691
PMID:27558654
-
Goto, S., Fujii, H., Kono, K., Watanabe, K., Nakai, K., Nishi, S., 2016. Clin Kidney J 9, 677–681.
PMCID: PMC5036911
PMID:27679714
-
Verheyen, N., Fahrleitner-Pammer, A., Pieske, B., Meinitzer, A., Belyavskiy, E., Wetzel, J., Gaksch, M., Grübler, M.R., Catena, C., Sechi, L.A., Van Ballegooijen, A.J., Brandenburg, V.M., Scharnagl, H., Perl, S., Brussee, H., März, W., Pilz, S., Tomaschitz, A., 2016. Journal of Hypertension 34, 1778–1786.
 Download biomedica product list 2018
Download biomedica product list 2018