LecChip™ – Lectin Microarray
-
Category number
GK-LECCHIP
-
Method
Lectin microarray detecting labeled carbohydrates/labeled cells
-
Sample type
Purified and crude samples, e.g. cell/tissue extracts, living cells, virus particles, cell culture supernatants, serum, urine
-
Sample volume
Purified/crude sample: 20 µl (50 µg/ml protein in PBS) Cells: 3×106 cells
-
Assay time
Cells: ~ 1 h
Purified/crude samples: overnight
-
Detection target
N- and O-glycans
-
Sensitivity
LOD: 100 pg/ml glycoprotein, 100 pM glycan, 103 cells
-
Regulatory status
Research use only.
Product Overview
Lectin microarrays using LecChip™ allows the highly sensitive analysis of carbohydrates in various sample types (purified and crude samples, e.g. cell/tissue extracts, living cells, virus particles, cell culture supernatants, serum, urine). Compared to MS-based methods, it requires less sample processing steps and is about 100 times more sensitive. Although it cannot identify glycan structures perfectly, it is a powerful system to identify differences of glycan structures like glycan isomers in a small amount of sample. Most importantly, it is possible to simultaneously gain data about O-glycans as well as N-glycans.
Analysis of LecChip™ requires an evanescent-field fluorescence excitation scanner like GlycoStation™ or GlycoLite™2200.
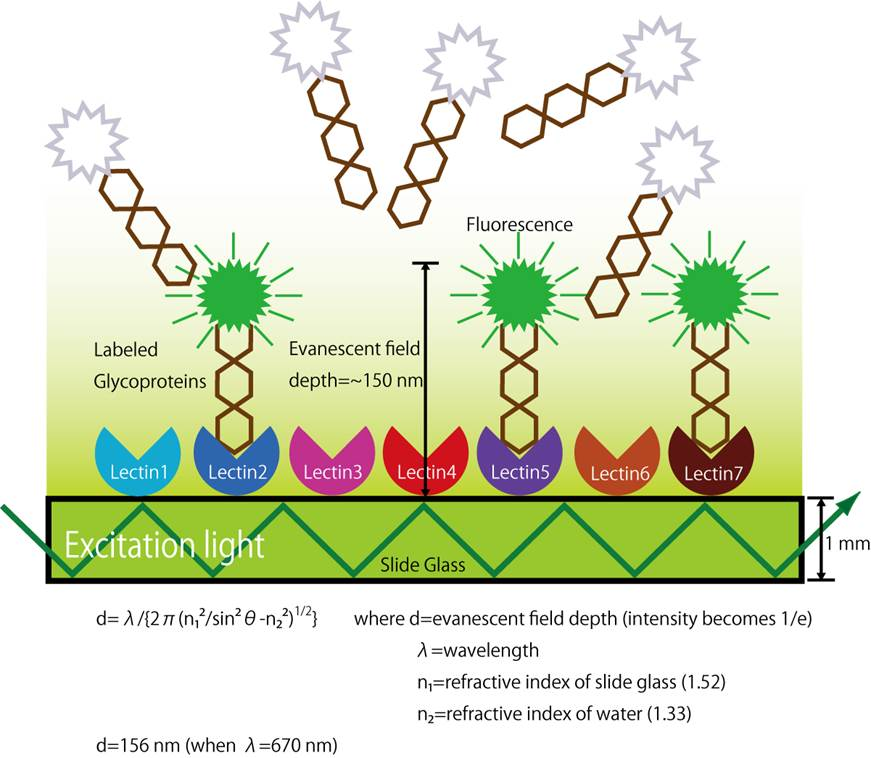
Principle of the LecChip™
The principle of glycan structure profiling analysis using LecChip™ is based on the lectin to glycan binding affinity. Lectins are glycan binding proteins and certain lectins will only bind to particular glycans. A LecChip™ contains 45 different lectins immobilized on a slide glass in an X-Y array. Cy3 labeled glycoproteins are applied to the LecChip™ surface which then bind to specific lectins on the chip. The LecChip™ is then analyzed with an evanescent-field fluorescence excitation scanner like GlycoStation™ or GlycoLite™2200 capable of generating an evanescent-field above the LecChip™ array. The Cy3 molecules attached to glycans that have successfully bound to lectins will fluoresce and the total LecChip™ fluorescence pattern can be captured with the scanner. Differential data analysis it best accomplished with the GlycoStation™ ToolsPro Ver.2.0 software.
The evanescent-field fluorescent excitation method used to excite the Cy3 marker on the glycans is an essential enabling technology as it allows for detection of very weak molecular interactions. Lectin-glycan interactions are known to be relatively weak compared to antigen-antibody and biotin-avidin interactions. If a washing process is applied to the LecChip™ to remove non-lectin binding redundant glycoproteins, many of the specifically bound glycans are also lost. Fortunately, evanescent-field fluorescence excitation allows for generation of lectin-glycan affinity data from unwashed samples by taking advantage of the higher signal strength of specifically bound glycans. The evanescent-field is formed on the surface of the LecChip™ when light enters into the slide glass from the sidewall and propagates through the glass. This is known as the principle of internal reflection mode. The evanescent field that is formed has a depth equal to the wavelength of the light and the field strength decreases exponentially with the distance away from the slide surface. Cy3-tagged glycans floating above the slide glass in a liquid phase exhibit a relatively low level of excitation as they are in the weakest part of the evanescent-field. In contrast, Cy3-tagged glycans that interact with the lectins located on the LecChip™ are contained in the stronger part of the evanescent field and are effectively excited. Hence, this technology allows monitoring of very weak molecular interactions directly from a liquid phase without washing of the sample.
Components
|
Contents |
Description |
Quantity |
|
LecChip™ |
Glass slide with 45 different immobilized lectins*, spotted in triplicates |
1 x chip |
* Lectins spotted on LecChips™:
|
No. |
Lectin (origin) |
Reported glycan selectivity |
|
1 |
LTL (Lotus tetragonolobus) |
Fucα1-3(Galβ1-4)GlcNAc (Lewis x), Fucα1-2Galβ1-4GlcNAc (H-type 2) |
|
2 |
PSA (Pisum sativum) |
Fucα1-6GlcNAc (Core Fuc) , α-Man |
|
3 |
LCA (Lens culinaris) |
Fucα1-6GlcNAc (Core Fuc), α-Man |
|
4 |
UEA-I (Ulex europaeus) |
Fucα1-2Galβ1-4GlcNAc (H-type 2) |
|
5 |
AOL (Aspergillus oryzae) |
Fucα1-6GlcNAc (Core Fuc), Fucα1-2Galβ1-4GlcNAc (H-type 2) |
|
6 |
AAL (Aleuria aurantia |
Fucα1-3(Galβ1-4)GlcNAc (Lewis x), Fucα1-6GlcNAc (Core Fuc) |
|
7 |
MAL_I (Maackia amurensis) |
Siaα2-3Galβ1-4GlcNAc |
|
8 |
SNA (Sambucus nigra) |
Siaα2-6Gal/GalNAc |
|
9 |
SSA (Sambucus sieboldiana) |
Siaα2-6Gal/GalNAc |
|
10 |
TJA-I (Trichosanthes japonica) |
Siaα2-6Gal/GalNAc, HSO3(-) -6Gal b1-4GlcNAc |
|
11 |
PHAL (Phaseolus vulgaris) |
tri/tetra-antennary complex-type N-glycan |
|
12 |
ECA (Erythrina cristagalli) |
Galβ1-4GlcNAc (up with increasing the number of terminal Gal), no affinity for fully sialylated N-type, fully agalactosylated N-type |
|
13 |
RCA120 (Ricinus communis) |
Galβ1-4GlcNAc (up with increasing the number of terminal Gal), Galb1-3Gal (weak), no affinity for agalactosylated N-type |
|
14 |
PHAE (Phaseolus vulgaris) |
bi-antennary complex-type N-glycan with outer Gal and bisecting GlcNAc, no affinity for fully sialylated N-type |
|
15 |
DSA (Datura stramonium) |
(GlcNAcβ1-4)n (Chitin), tri/tetra-antennary N-glycan |
|
16 |
GSL-II (Griffonia simplicifolia) |
agalactosylated tri/tetra antennary glycans, GlcNAc, no affinity for fully galactosylated or sialylated N-type |
|
17 |
NPA (Narcissus pseudonarcissus) |
High-Mannose including Manα1-6Man |
|
18 |
ConA (Canavalia ensiformis) |
High-Mannose including Manα1-6(Manα1-3)Man |
|
19 |
GNA (Galanthus nivalis) |
High-Mannose including Manα1-3Man |
|
20 |
HHL (Hippeastrum hybrid) |
High-Mannose including Manα1-3Man or Manα1-6Man |
|
21 |
ACG (mushroom, Agrocybe cylindracea) |
Gal b1-3Gal, Siaα2-3Galβ1-4GlcNAc |
|
22 |
TxLCI (Tulipa gesneriana) |
Manα1-3(Manα1-6)Man, bi/tri-antennary complex-type N-glycan, GalNAc |
|
23 |
BPL (Bauhinia purpurea) |
Galβ1-3GalNAc (up with Lewis x, down with Core Fuc), GalNAc |
|
24 |
TJA-II (Tanthes japonica) |
Fucα1-2Galβ1-> or GalNAcβ1-> groups at their non-reducing terminals |
|
25 |
EEL (Euonymus europaeus) |
Gala1-3Galb1-4GlcNAc, Fuca1-2Galb1-3GlcNAc (H antigen) |
|
26 |
ABA (fungus, Agaricus bisporus) |
Galβ1-3GalNAc, GlcNAc |
|
27 |
LEL tomato, Lycopersicon esculentum) |
(GlcNAcβ1-4)n (Chitin), (Galb1-4GlcNAc)n (polylactosamine) |
|
28 |
STL (potato, Solanum tuberosum) |
(GlcNAcβ1-4)n (Chitin) oligosaccharide containing GlcNAc and MurNAc |
|
29 |
UDA (Urtica dioica) |
GlcNAcβ1-4GlcNAc (Chitin), High-Mannose (3 to High, up with increasing the number of Man) |
|
30 |
PWM (pokeweed, Phytolacca Americana) |
(GlcNAcβ1-4)n (Chitin) |
|
31 |
Jacalin (Artocarpus integrifolia) |
GlcNAcb1-3GalNAc (Core3), Siaa2-3Galb1-3GalNAc (sialyl T), Galb1-3GalNAc (T-antigen), a-GalNAc (Tn-antigen) |
|
32 |
PNA (peanut, Arachis hypogaea) |
Galβ1-3GalNAc |
|
33 |
WFA (Wisteria floribunda) |
GalNAcβ1-4GlcNAc (LacdiNAc), Galβ1-3(-6)GalNAc |
|
34 |
ACA (Amaranthus caudatus) |
Galβ1-3GalNAc (T-antigen), Siaa2-3Galb1- GalNAc (sialyl T) |
|
35 |
MPA (Maclura pomifera) |
a-GalNAc (Tn-antigen), Galβ1-3GalNAc (T-antigen) |
|
36 |
HPA (snail, Helix pomatia) |
α-GalNAc |
|
37 |
VVA (Vicia villosa) |
GalNAcb1-4Gal, GalNAcb1-3Gal, a-GalNAc, |
|
38 |
DBA (Dolichos biflorus) |
Blood group A, GalNAcα1-3GalNAc, GalNAcb1-4(Siaa2-3)Galb1-4Glc (GM2) |
|
39 |
SBA (soybean, Dolichos biflorus) |
α- or β-linked GalNAc, Gala1-4Gal-Glc |
|
40 |
Calsepa (Calystegia sepium) |
Galactosylated bianntenary N-type with bisecting GlcNAc (galacto > agalacto, down with Core Fuc), High-Mannose (Man2–6) |
|
41 |
PTL-I (Psophocarpus tetragonolobus) |
α-GalNAc, Gala1-3(Fuca1-2)Gal (B-antigen) |
|
42 |
MAH (Maackia amurensis) |
Siaα2-3Galβ1-3(Siaα2-6)GalNAc (disialyl-T) |
|
43 |
WGA (wheat germ, Triticum aestivum) |
(GlcNAcβ1-4)n (Chitin), Hybrid type N-glycan, Sia |
|
44 |
GSL-I A4 (Griffonia simplicifolia) |
α-GalNAc |
|
45 |
GSL-I B4 (Griffonia simplicifolia) |
α-Gal |
Assay Protocols
Background
Glycosylation is a common posttranslational modification of proteins and influences parameters like protein folding, targeting, ligand recognition, stability, immunogenicity, biological activity, etc. Glycans play a significant biological role in mainly four medical areas: cancer, immunology, infectious diseases and regenerative medicine. Here, a quick, simple and highly sensitive method for glycan profile analysis can accelerate the development of advanced biomarkers, therapeutic proteins, probiotic methodologies as well as the characterization of stem cell (iPS, ES, and MSC).
Applications
-
Detection of Changes in Glycosylation Pattern
-
Supporting Quality Control Activities (Batch-to-Batch Variation)
-
Comparability Testing of Biosimilars
-
Monitoring of Biologicals (e.g. Protein Drugs)
-
Host Cell Characterization
-
Glyco-Biomarker Discovery
-
Characterization of Stem Cells & Differentiated Cells
-
Advanced Histochemistry (e.g. Marker Definition)
-
Production Process Optimization (IPC testing)
- Lectin microarray analyses reveal host cell-specific glycan profiles of the hemagglutinins of influenza A viruses.
Hiono, T., Matsuda, A., Wagatsuma, T., Okamatsu, M., Sakoda, Y., Kuno, A., 2019. Virology 527, 132–140.
- Lectin microarray analyses reveal host cell-specific glycan profiles of the hemagglutinins of influenza A viruses.Analysis of a lectin microarray identifies altered sialylation of mouse serum glycoproteins induced by whole-body radiation exposure.
vIizuka, D., Izumi, S., Suzuki, F., Kamiya, K., 2018. J Radiat Res.
- Characteristic glycopeptides associated with extreme human longevity identified through plasma glycoproteomics.
Miura, Y., Hashii, N., Ohta, Y., Itakura, Y., Tsumoto, H., Suzuki, J., Takakura, D., Abe, Y., Arai, Y., Toyoda, M., Kawasaki, N., Hirose, N., Endo, T., 2018. Biochimica et Biophysica Acta (BBA) - General Subjects 1862, 1462–1471.
- Glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis.
Noda, M., Okayama, H., Tachibana, K., Sakamoto, W., Saito, K., Min, A.K.T., Ashizawa, M., Nakajima, T., Aoto, K., Momma, T., Katakura, K., Ohki, S., Kono, K., 2018. Clin Cancer Res clincanres.3533.2017.
PMID:29844132
- Possible role of sialylation of retinal protein glycans in the regulation of electroretinogram response in mice.
Ahuja, S., 2017. Int J Ophthalmol 10, 1217–1222. PMCID: PMC5554838
PMID:28861345
- Assessment of tumor characteristics based on glycoform analysis of membrane-tethered MUC1.
Matsuda, A., Higashi, M., Nakagawa, T., Yokoyama, S., Kuno, A., Yonezawa, S., Narimatsu, H., 2017. Laboratory Investigation 97, 1103–1113.
- Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC.
Roucka, M., Zimmermann, K., Fido, M., Nechansky, A., 2017. Microarrays 6, 1.
- Glycan profiling analysis using evanescent-field fluorescence-assisted lectin array: Importance of sugar recognition for cellular uptake of exosomes from mesenchymal stem cells.
Shimoda, A., Tahara, Y., Sawada, S., Sasaki, Y., Akiyoshi, K., 2017. Biochemical and Biophysical Research Communications 491, 701–707.
- Glycan profiling using formalin-fixed, paraffin-embedded tissues: Hippeastrum hybrid lectin is a sensitive biomarker for squamous cell carcinoma of the uterine cervix.
Tozawa‐Ono, A., Kubota, M., Honma, C., Nakagawa, Y., Yokomichi, N., Yoshioka, N., Tsuda, C., Ohara, T., Koizumi, H., Suzuki, N., 2017. Journal of Obstetrics and Gynaecology Research 43, 1326–1334.
- Lectin array and glycogene expression analyses of ovarian cancer cell line A2780 and its cisplatin-resistant derivate cell line A2780-cp.
Zhao, R., Qin, W., Qin, R., Han, J., Li, C., Wang, Y., Xu, C., 2017. Clinical Proteomics 14, 20.
- Aberrant Glycosylation in the Left Ventricle and Plasma of Rats with Cardiac Hypertrophy and Heart Failure.
Nagai-Okatani, C., Minamino, N., 2016. PLOS ONE 11, e0150210.
- Alteration of matrix metalloproteinase-3 O-glycan structure as a biomarker for disease activity of rheumatoid arthritis.
Takeshita, M., Kuno, A., Suzuki, K., Matsuda, A., Shimazaki, H., Nakagawa, T., Otomo, Y., Kabe, Y., Suematsu, M., Narimatsu, H., Takeuchi, T., 2016. Arthritis Research & Therapy 18, 112.
- Fucosylation is associated with the malignant transformation of intraductal papillary mucinous neoplasms: a lectin microarray-based study.
Watanabe, K., Ohta, M., Yada, K., Komori, Y., Iwashita, Y., Kashima, K., Inomata, M., 2016. Surg Today 46, 1217–1223.
- Integrated glycomic analysis of ovarian cancer side population cells.
Zhao, R., Liu, X., Wang, Y., Jie, X., Qin, R., Qin, W., Zhang, M., Tai, H., Yang, C., Li, L., Peng, P., Shao, M., Zhang, X., Wu, H., Ruan, Y., Xu, C., Ren, S., Gu, J., 2016. Clinical Proteomics 13, 32.
- Decreased expression of Bauhinia purpurea lectin is a predictor of gastric cancer recurrence.
Futsukaichi, T., Etoh, T., Nakajima, K., Daa, T., Shiroshita, H., Shiraishi, N., Kitano, S., Inomata, M., 2015. Surg Today 45, 1299–1306.
- Development and Applications of the Lectin Microarray.
Hirabayashi, J., Kuno, A., Tateno, H., 2015, in: Gerardy-Schahn, R., Delannoy, P., von Itzstein, M. (Eds.), SialoGlyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans, Topics in Current Chemistry. Springer International Publishing, Cham, pp. 105–124.
- Monoclonal Antibody LpMab-9 Recognizes O-glycosylated N-Terminus of Human Podoplanin.
Kaneko, M.K., Oki, H., Hozumi, Y., Liu, X., Ogasawara, S., Takagi, M., Goto, K., Kato, Y., 2015. Monoclonal Antibodies in Immunodiagnosis and Immunotherapy 34, 310–317.
- Establishment of new predictive markers for distant recurrence of colorectal cancer using lectin microarray analysis.
Nakajima, K., Inomata, M., Iha, H., Hiratsuka, T., Etoh, T., Shiraishi, N., Kashima, K., Kitano, S., 2015. Cancer Medicine 4, 293–302.
- Development of Monoclonal Antibody LpMab-10 Recognizing Non-glycosylated PLAG1/2 Domain Including Thr34 of Human Podoplanin.
Ogasawara, S., Oki, H., Kaneko, M.K., Hozumi, Y., Liu, X., Honma, R., Fujii, Y., Nakamura, T., Goto, K., Takagi, M., Kato, Y., 2015. Monoclonal Antibodies in Immunodiagnosis and Immunotherapy 34, 318–326.
- Dynamic regulation of innate immune responses in Drosophila by Senju-mediated glycosylation.
Yamamoto-Hino, M., Muraoka, M., Kondo, S., Ueda, R., Okano, H., Goto, S., 2015. PNAS 201424514.
PMID:25901322
- Differential Glycan Analysis of an Endogenous Glycoprotein: Toward Clinical Implementation—From Sample Pretreatment to Data Standardization.
Kuno, A., Matsuda, A., Unno, S., Tan, B., Hirabayashi, J., Narimatsu, H., 2014, in: Hirabayashi, J. (Ed.), Lectins: Methods and Protocols, Methods in Molecular Biology. Springer New York, New York, NY, pp. 265–285.
- Lectin Microarray Profiling and Relative Quantification of Glycome Associated With Proteins of Neonatal wt and rd1 Mice Retinae.
Ahuja, S., 2013. Invest. Ophthalmol. Vis. Sci. 54, 3272–3280.
- Binding sugars: from natural lectins to synthetic receptors and engineered neolectins.
Arnaud, J., Audfray, A., Imberty, A., 2013. Chem. Soc. Rev. 42, 4798–4813.
- β-mannosyl linkages inhibit CAWS arteritis by negatively regulating dectin-2-dependent signaling in spleen and dendritic cells.
Hirata, N., Ishibashi, K., Sato, W., Nagi-Miura, N., Adachi, Y., Ohta, S., Ohno, N., 2013. Immunopharmacology and Immunotoxicology 35, 594–604.
- Urinary Fetuin-A Is a Novel Marker for Diabetic Nephropathy in Type 2 Diabetes Identified by Lectin Microarray.
Inoue, K., Wada, J., Eguchi, J., Nakatsuka, A., Teshigawara, S., Murakami, K., Ogawa, D., Terami, T., Katayama, A., Tone, A., Iseda, I., Hida, K., Yamada, M., Ogawa, T., Makino, H., 2013. PLOS ONE 8, e77118.
- Large-scale cell production of stem cells for clinical application using the automated cell processing machine.
Kami, D., Watakabe, K., Yamazaki-Inoue, M., Minami, K., Kitani, T., Itakura, Y., Toyoda, M., Sakurai, T., Umezawa, A., Gojo, S., 2013. BMC Biotechnology 13, 102.
- Glycoproteomics-based cancer marker discovery adopting dual enrichment with Wisteria floribunda agglutinin for high specific glyco-diagnosis of cholangiocarcinoma.
Matsuda, A., Kuno, A., Matsuzaki, H., Kawamoto, T., Shikanai, T., Nakanuma, Y., Yamamoto, M., Ohkohchi, N., Ikehara, Y., Shoda, J., Hirabayashi, J., Narimatsu, H., 2013. Journal of Proteomics 85, 1–11.
- Multilectin-assisted fractionation for improved single-dot tissue glycome profiling in clinical glycoproteomics.
Tan, B., Matsuda, A., Zhang, Y., Kuno, A., Narimatsu, H., 2013. Mol. BioSyst. 10, 201–205.
- Lectin Array-Based Strategies for Identifying Metastasis-Associated Changes in Glycosylation.
Fry, S., Afrough, B., Leathem, A., Dwek, M., 2012, in: Dwek, M., Brooks, S.A., Schumacher, U. (Eds.), Metastasis Research Protocols, Methods in Molecular Biology. Humana Press, Totowa, NJ, pp. 267–272.
- LecT-Hepa: A triplex lectin–antibody sandwich immunoassay for estimating the progression dynamics of liver fibrosis assisted by a bedside clinical chemistry analyzer and an automated pretreatment machine.
Kuno, A., Ikehara, Y., Tanaka, Y., Saito, K., Ito, K., Tsuruno, C., Nagai, S., Takahama, Y., Mizokami, M., Hirabayashi, J., Narimatsu, H., 2011. Clinica Chimica Acta 412, 1767–1772.
- Chapter Seven - Differential Glycan Profiling by Lectin Microarray Targeting Tissue Specimens.
Kuno, A., Matsuda, A., Ikehara, Y., Narimatsu, H., Hirabayashi, J., 2010, in: Fukuda, M. (Ed.), Methods in Enzymology, Glycomics. Academic Press, pp. 165–179.
- Transient expression of an IL-23R extracellular domain Fc fusion protein in CHO vs. HEK cells results in improved plasma exposure.
Suen, K.F., Turner, M.S., Gao, F., Liu, B., Althage, A., Slavin, A., Ou, W., Zuo, E., Eckart, M., Ogawa, T., Yamada, M., Tuntland, T., Harris, J.L., Trauger, J.W., 2010. Protein Expression and Purification 71, 96–102.
- Microarray Methods and Protocols.
Matson, R.S., 2009. CRC Press
- Microarray Methods and Protocols.
Matson, R.S., 2009. CRC Press
 Download biomedica product list 2018
Download biomedica product list 2018